Virginie Callot
Monitoring morphometric drift in lifelong learning segmentation of the spinal cord
May 02, 2025Abstract:Morphometric measures derived from spinal cord segmentations can serve as diagnostic and prognostic biomarkers in neurological diseases and injuries affecting the spinal cord. While robust, automatic segmentation methods to a wide variety of contrasts and pathologies have been developed over the past few years, whether their predictions are stable as the model is updated using new datasets has not been assessed. This is particularly important for deriving normative values from healthy participants. In this study, we present a spinal cord segmentation model trained on a multisite $(n=75)$ dataset, including 9 different MRI contrasts and several spinal cord pathologies. We also introduce a lifelong learning framework to automatically monitor the morphometric drift as the model is updated using additional datasets. The framework is triggered by an automatic GitHub Actions workflow every time a new model is created, recording the morphometric values derived from the model's predictions over time. As a real-world application of the proposed framework, we employed the spinal cord segmentation model to update a recently-introduced normative database of healthy participants containing commonly used measures of spinal cord morphometry. Results showed that: (i) our model outperforms previous versions and pathology-specific models on challenging lumbar spinal cord cases, achieving an average Dice score of $0.95 \pm 0.03$; (ii) the automatic workflow for monitoring morphometric drift provides a quick feedback loop for developing future segmentation models; and (iii) the scaling factor required to update the database of morphometric measures is nearly constant among slices across the given vertebral levels, showing minimum drift between the current and previous versions of the model monitored by the framework. The model is freely available in Spinal Cord Toolbox v7.0.
Segmentation of Multiple Sclerosis Lesions across Hospitals: Learn Continually or Train from Scratch?
Oct 27, 2022



Abstract:Segmentation of Multiple Sclerosis (MS) lesions is a challenging problem. Several deep-learning-based methods have been proposed in recent years. However, most methods tend to be static, that is, a single model trained on a large, specialized dataset, which does not generalize well. Instead, the model should learn across datasets arriving sequentially from different hospitals by building upon the characteristics of lesions in a continual manner. In this regard, we explore experience replay, a well-known continual learning method, in the context of MS lesion segmentation across multi-contrast data from 8 different hospitals. Our experiments show that replay is able to achieve positive backward transfer and reduce catastrophic forgetting compared to sequential fine-tuning. Furthermore, replay outperforms the multi-domain training, thereby emerging as a promising solution for the segmentation of MS lesions. The code is available at this link: https://github.com/naga-karthik/continual-learning-ms
2D Multi-Class Model for Gray and White Matter Segmentation of the Cervical Spinal Cord at 7T
Oct 13, 2021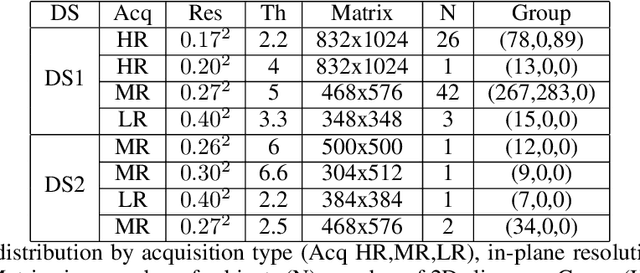
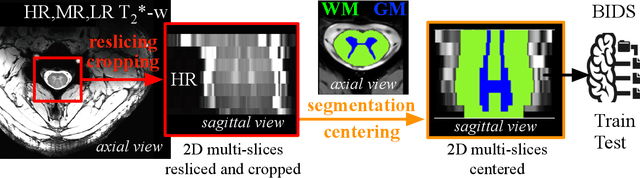
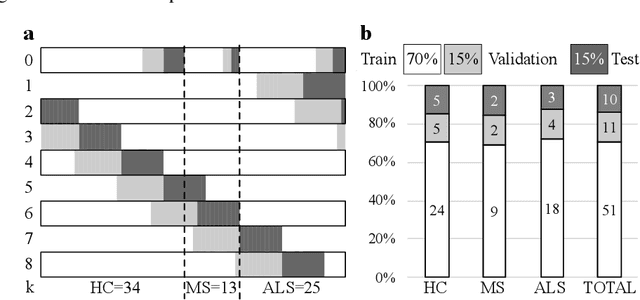
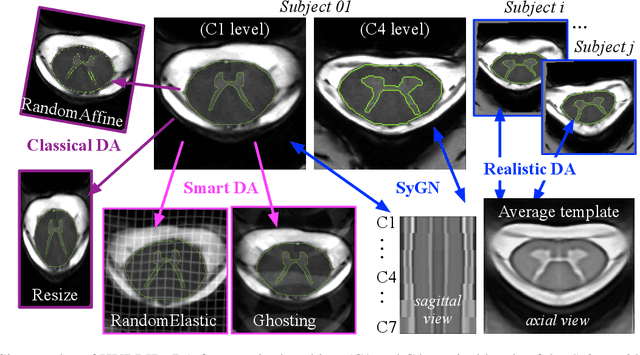
Abstract:The spinal cord (SC), which conveys information between the brain and the peripheral nervous system, plays a key role in various neurological disorders such as multiple sclerosis (MS) and amyotrophic lateral sclerosis (ALS), in which both gray matter (GM) and white matter (WM) may be impaired. While automated methods for WM/GM segmentation are now largely available, these techniques, developed for conventional systems (3T or lower) do not necessarily perform well on 7T MRI data, which feature finer details, contrasts, but also different artifacts or signal dropout. The primary goal of this study is thus to propose a new deep learning model that allows robust SC/GM multi-class segmentation based on ultra-high resolution 7T T2*-w MR images. The second objective is to highlight the relevance of implementing a specific data augmentation (DA) strategy, in particular to generate a generic model that could be used for multi-center studies at 7T.
Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks
Sep 11, 2018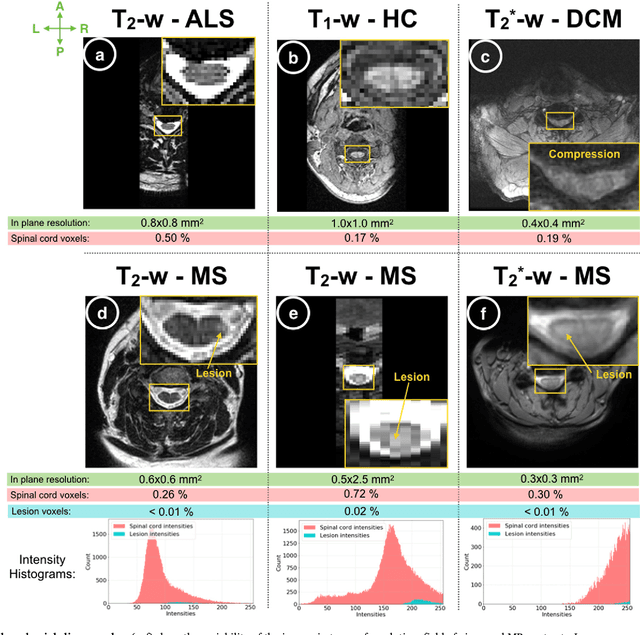
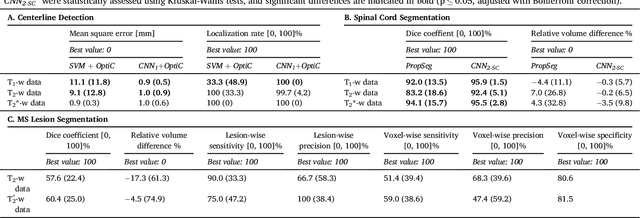
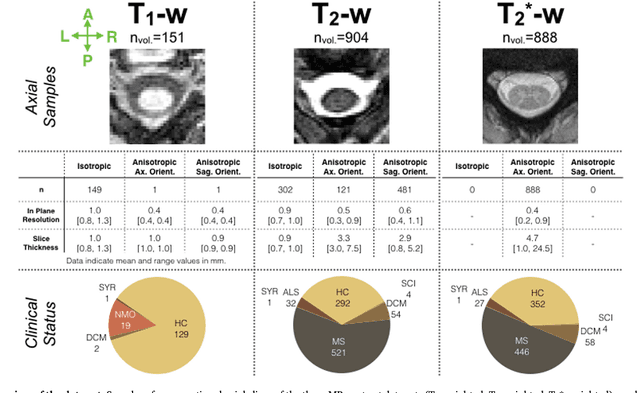
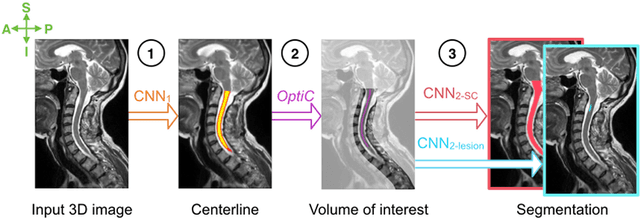
Abstract:The spinal cord is frequently affected by atrophy and/or lesions in multiple sclerosis (MS) patients. Segmentation of the spinal cord and lesions from MRI data provides measures of damage, which are key criteria for the diagnosis, prognosis, and longitudinal monitoring in MS. Automating this operation eliminates inter-rater variability and increases the efficiency of large-throughput analysis pipelines. Robust and reliable segmentation across multi-site spinal cord data is challenging because of the large variability related to acquisition parameters and image artifacts. The goal of this study was to develop a fully-automatic framework, robust to variability in both image parameters and clinical condition, for segmentation of the spinal cord and intramedullary MS lesions from conventional MRI data. Scans of 1,042 subjects (459 healthy controls, 471 MS patients, and 112 with other spinal pathologies) were included in this multi-site study (n=30). Data spanned three contrasts (T1-, T2-, and T2*-weighted) for a total of 1,943 volumes. The proposed cord and lesion automatic segmentation approach is based on a sequence of two Convolutional Neural Networks (CNNs). To deal with the very small proportion of spinal cord and/or lesion voxels compared to the rest of the volume, a first CNN with 2D dilated convolutions detects the spinal cord centerline, followed by a second CNN with 3D convolutions that segments the spinal cord and/or lesions. When compared against manual segmentation, our CNN-based approach showed a median Dice of 95% vs. 88% for PropSeg, a state-of-the-art spinal cord segmentation method. Regarding lesion segmentation on MS data, our framework provided a Dice of 60%, a relative volume difference of -15%, and a lesion-wise detection sensitivity and precision of 83% and 77%, respectively. The proposed framework is open-source and readily available in the Spinal Cord Toolbox.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge